Plotting temporal residuals
2022-06-17
Source:vignettes/a03_temporal_residuals.Rmd
a03_temporal_residuals.RmdHere we will use the vista package to plot temporal residuals from spatiotemporal models
GAM with spatial smooths by year
We’ll use a GAM for demonstration purposes, but any other package
randomForest, sdmTMB, etc. could be used
instead.
Predictions through time
The pred_time function displays the range of predictions
across time slices.
pred_time(df = pcod, time = "year")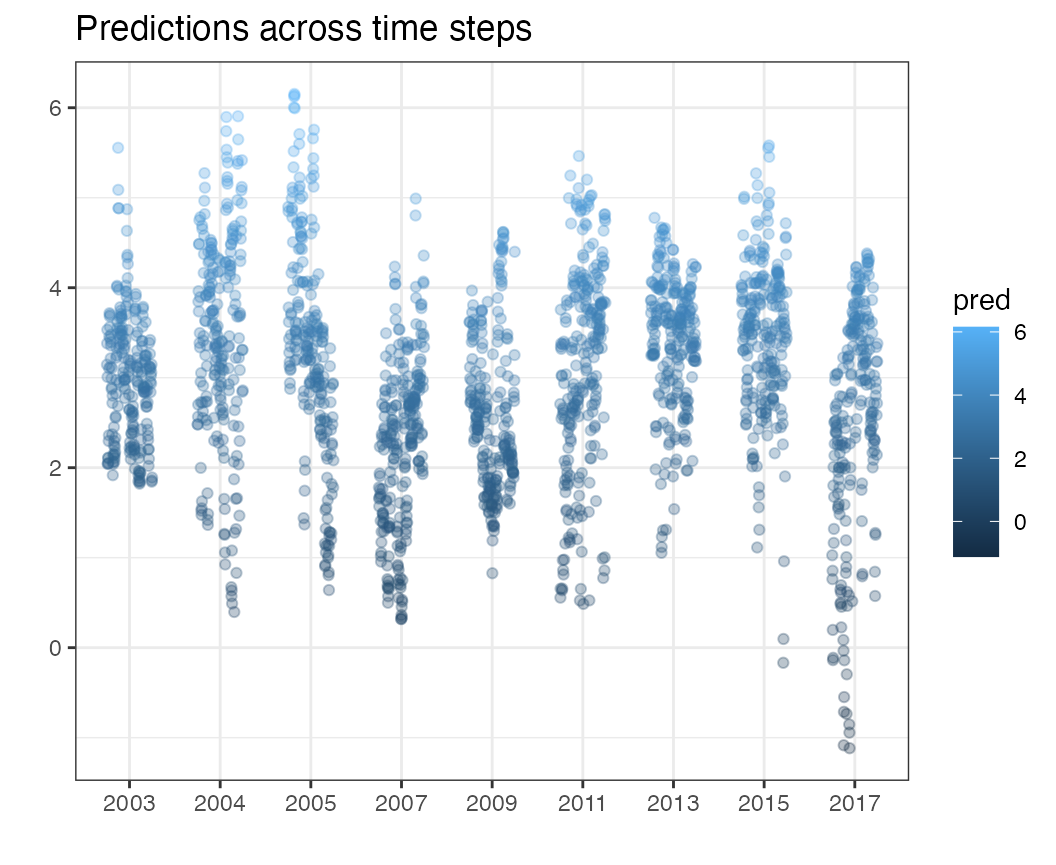
Residuals through time
Similarly, the resid_time function is designed to plot
residuals across time slices
resid_time(df = pcod, time = "year")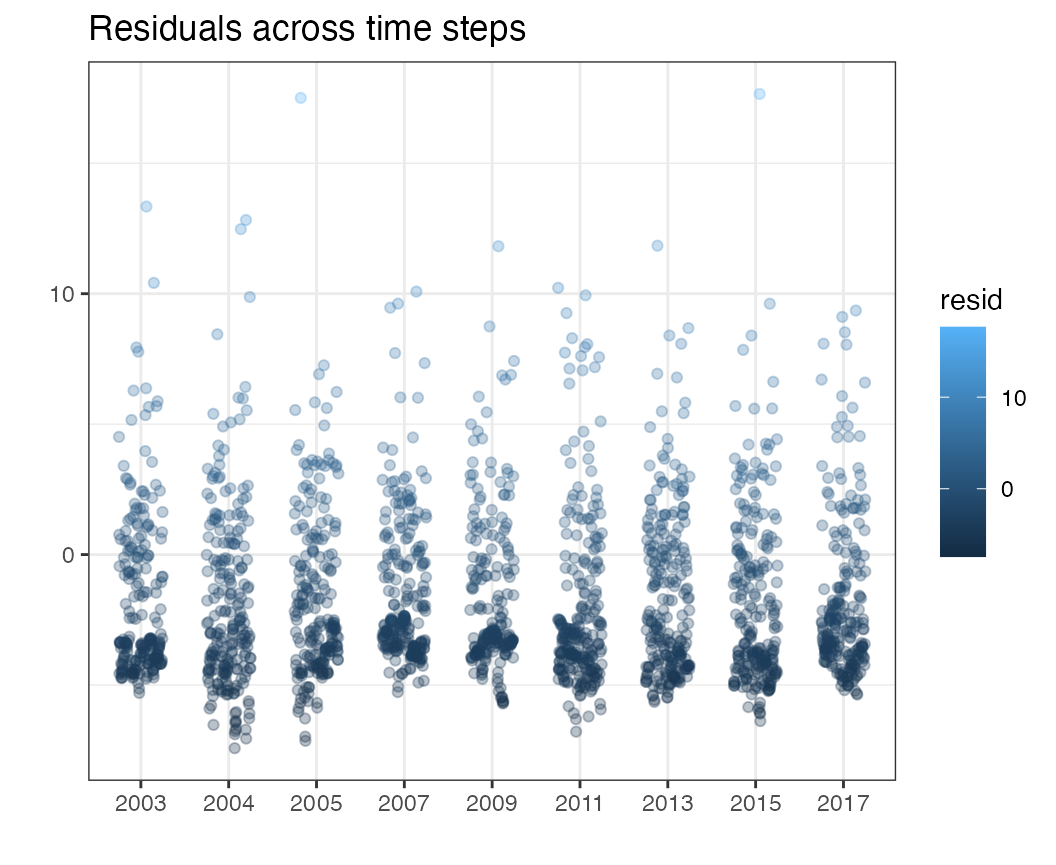
Homogeneity of residuals through time
In some cases, the variability of a spatial process may be
non-stationary and increasing or decreasing over time. We can visualize
this using the sd_resid_time function, where ideally there
is no trend or outlier.
sd_resid_time(df = pcod, time = "year")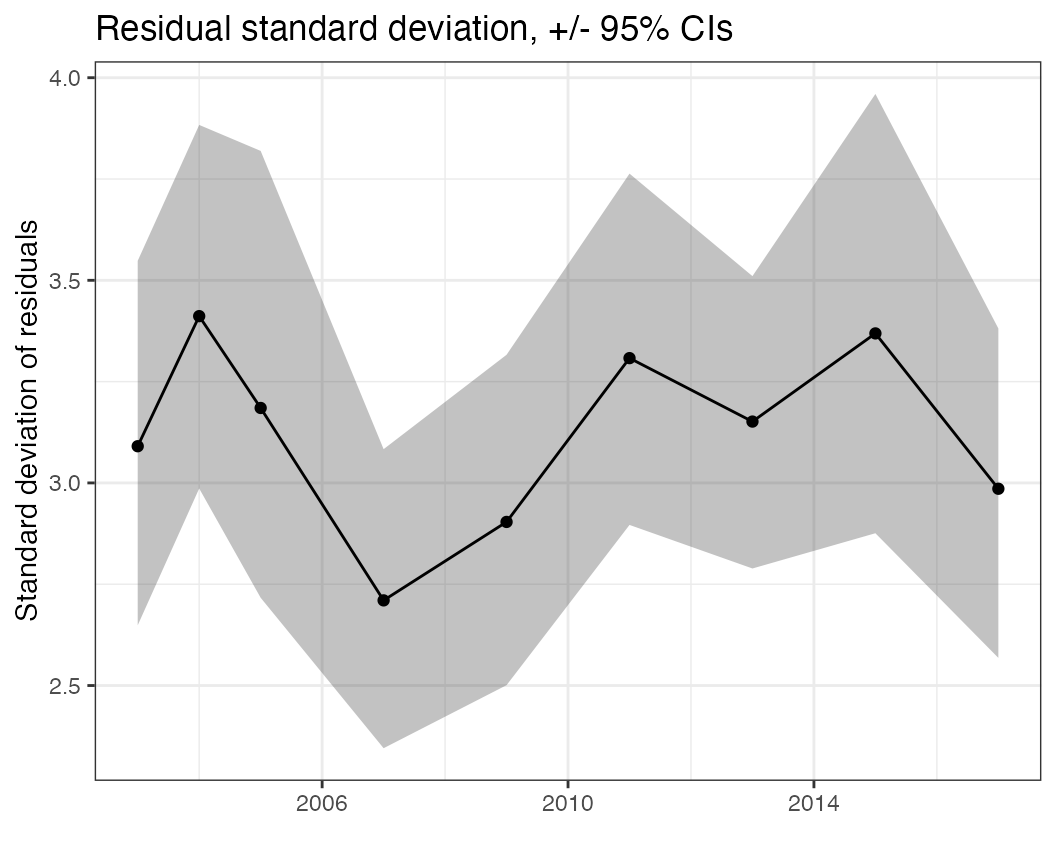
QQ plot temporally
The qq function is also useful in displaying the
quantile residuals (via a call to stats::qqnorm). By
default this will be faceted by time, but faceting can be turned off
with by_time = FALSE.
qq(pcod, time = "year")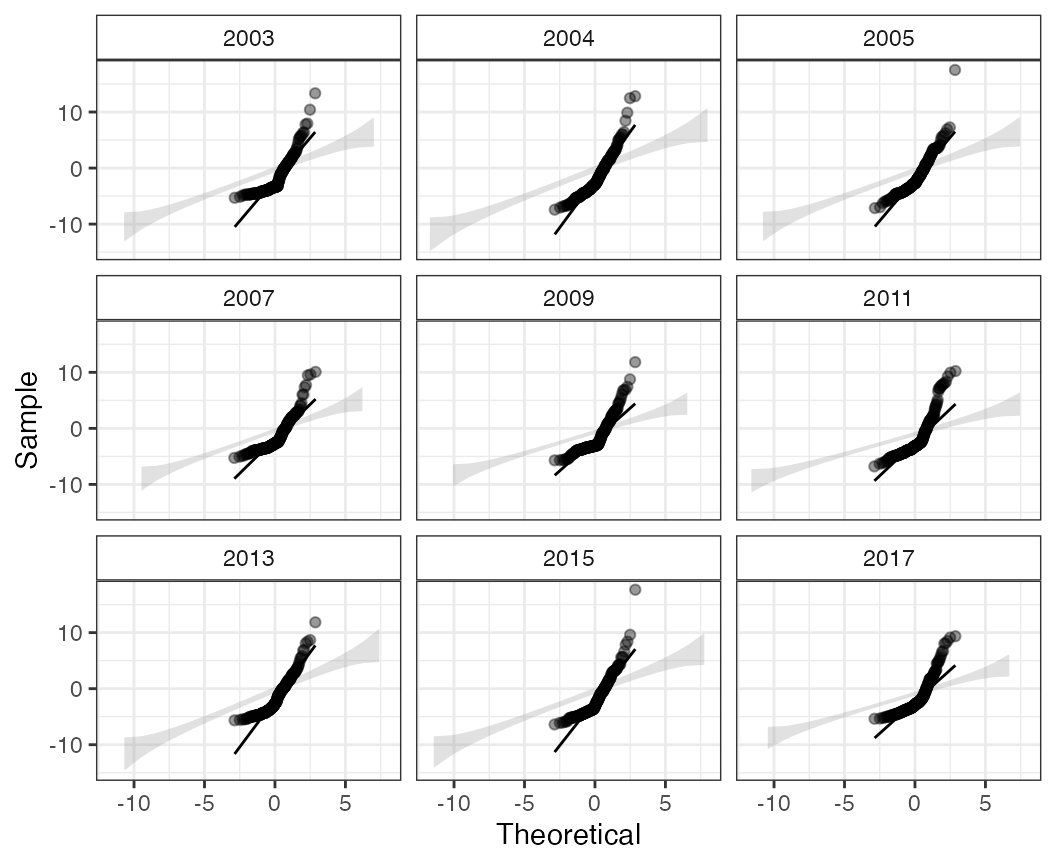
Scale - location plots
The scale_loc function displays scale - location plots,
which may be useful at diagnosing trends in residuals vs predicted
values,
scale_loc(df = pcod, time="year")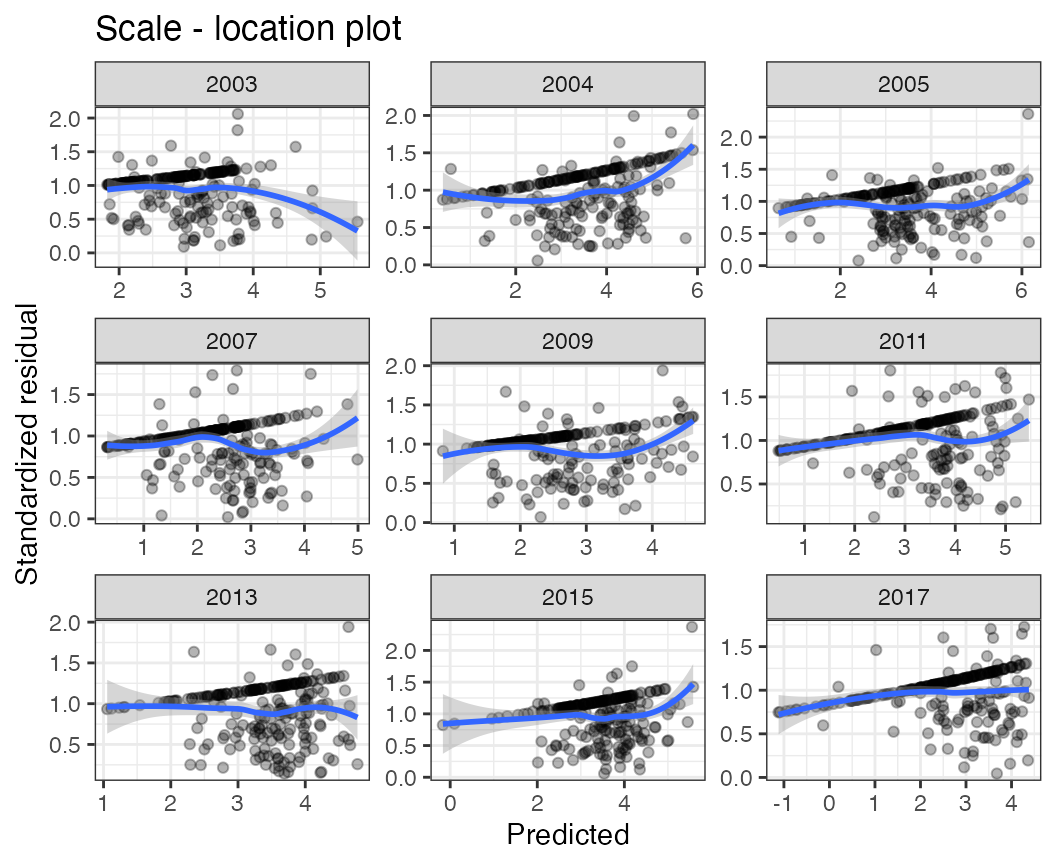
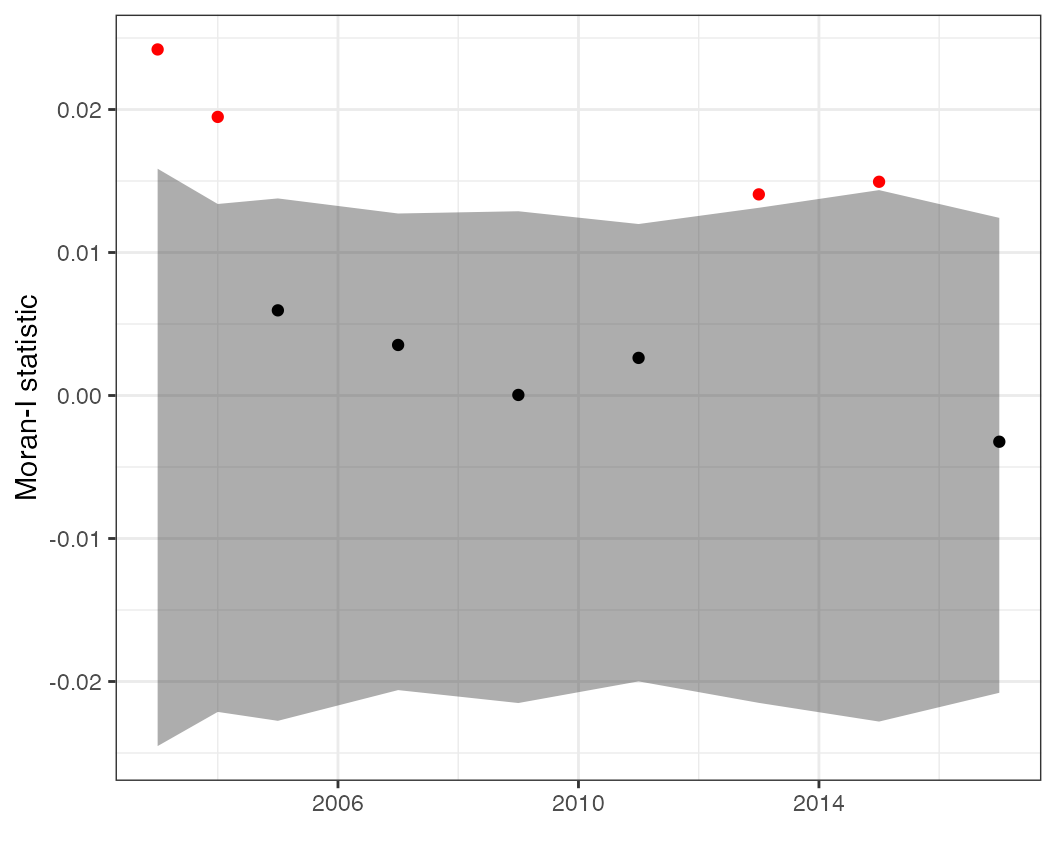
Moran’s I statistics
We can calculate lots of variants of Moran’s I. The
moran_ts plot is useful for displaying time series of the
Moran-I statistic, calculated separately for each time slice. This can
be done on predicted values,
moran_ts(df = pcod, time = "year", response = "pred")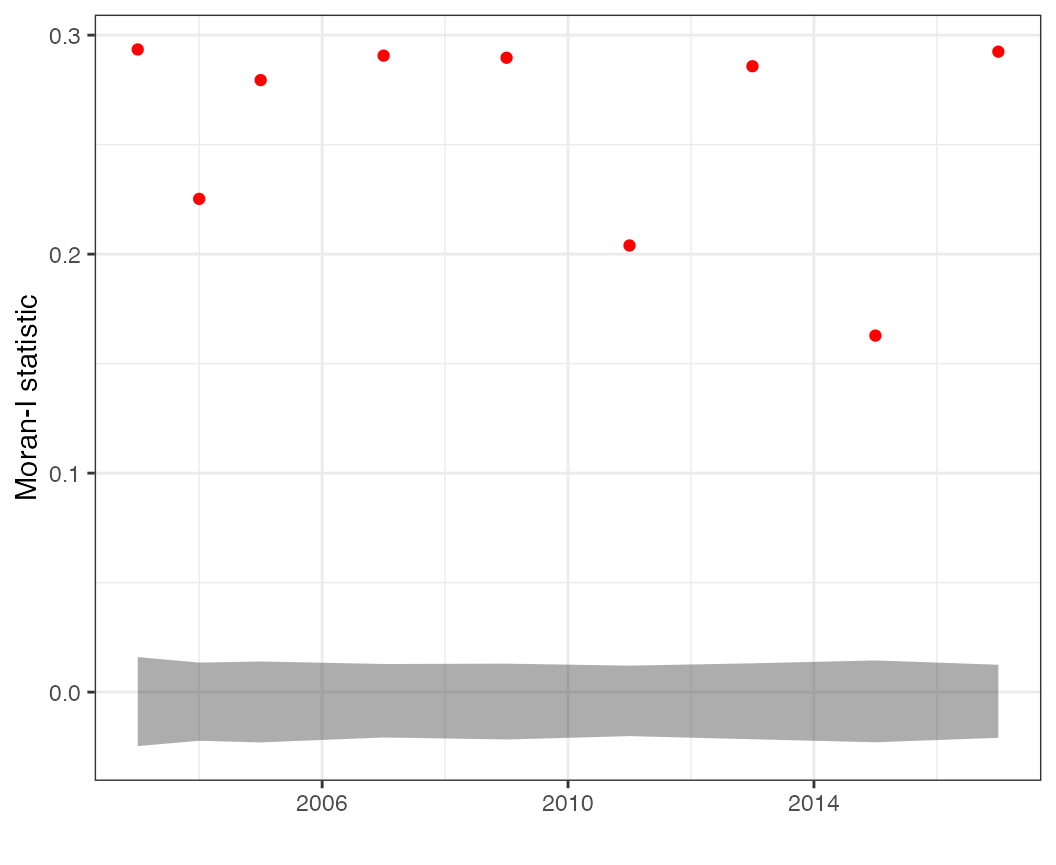
But if a goal is evaluating whether a particular model removes spatial autocorrelation, it may be more useful to calculate this on the residuals,
moran_ts(df = pcod, time = "year", response = "resid")